Future Horizons:
10-yearhorizon
Past ecosystems are reconstructed through AI and environmental DNA
AI enables identification of the most common flora and fauna specimens, such as pollen and teeth. Environmental DNA from sediments and other sources is routinely used to reconstruct past ecosystems. Researchers reconstruct past food webs using a combination of isotope analyses, tooth wear and other tools.
25-yearhorizon
Drivers of past extinctions brought to light
Animals, plants and other organisms can be identified from their physical remains, from DNA and from proteomics. A particularly valuable technique developed in the last decade is ZooMS, which involves extracting protein from tiny fragments of bone in order to identify species.13 This has enabled the identification of animal foods in ancient hominin sites14 and revealed hominins’ interactions with other large animals.15,16
These new techniques have also enabled us to revise our understanding of the environments in which hominins lived17 and to tease out the interactions between humans and their environments. There is growing evidence that our ancestors and relatives had negative impacts,18 contributing to the extinctions of many species, especially large “megafauna”.19Environmental shifts also impacted hominins and their evolution,20,21 even driving the evolution of new species22
The reconstruction of ancient pathogens relies on the ability to separate pathogen DNA from that of its human host.23 Combined with existing techniques, such as identifying traces of disease on skeletons and burial patterns,24 such studies help us understand the ecology and evolution of diseases. They also reveal how societies and technologies, such as the invention of farming, have impacted the spread of pathogens.25 In addition, they offer support for the contention that human health is inextricably connected with ecosystems, as suggested in the One Health model.26
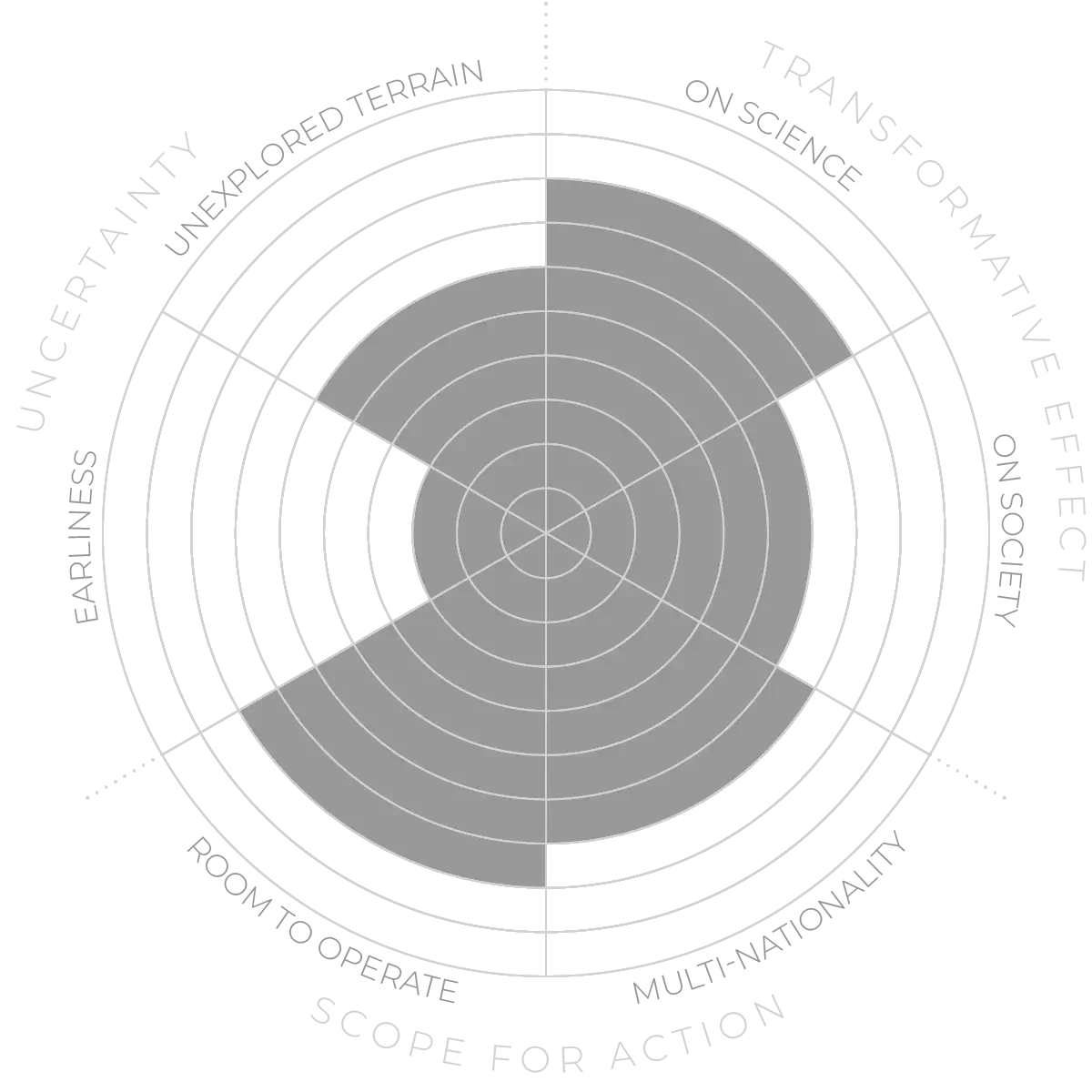
Molecular-level analysis of ancient non-human remains - Anticipation Scores
The Anticipation Potential of a research field is determined by the capacity for impactful action in the present, considering possible future transformative breakthroughs in a field over a 25-year outlook. A field with a high Anticipation Potential, therefore, combines the potential range of future transformative possibilities engendered by a research area with a wide field of opportunities for action in the present. We asked researchers in the field to anticipate:
- The uncertainty related to future science breakthroughs in the field
- The transformative effect anticipated breakthroughs may have on research and society
- The scope for action in the present in relation to anticipated breakthroughs.
This chart represents a summary of their responses to each of these elements, which when combined, provide the Anticipation Potential for the topic. See methodology for more information.